Architecture
Chunkwise consists of two major subsystems:
-
Experimentation Platform - supports interactive chunk visualization and retrieval-based evaluation. It adopts a microservice architecture consisting of four components:
- Chunking Service
- Evaluation Service
- Backend Server
- Client
-
Data Ingestion Pipeline - applies a selected chunking strategy to the user’s entire corpus and stores embeddings in a vector database for downstream RAG applications.
These two subsystems are unified through the backend server, which coordinates both experimentation workflows and pipeline deployments.
System Architecture
Section titled “System Architecture”Chunking Service
Section titled “Chunking Service”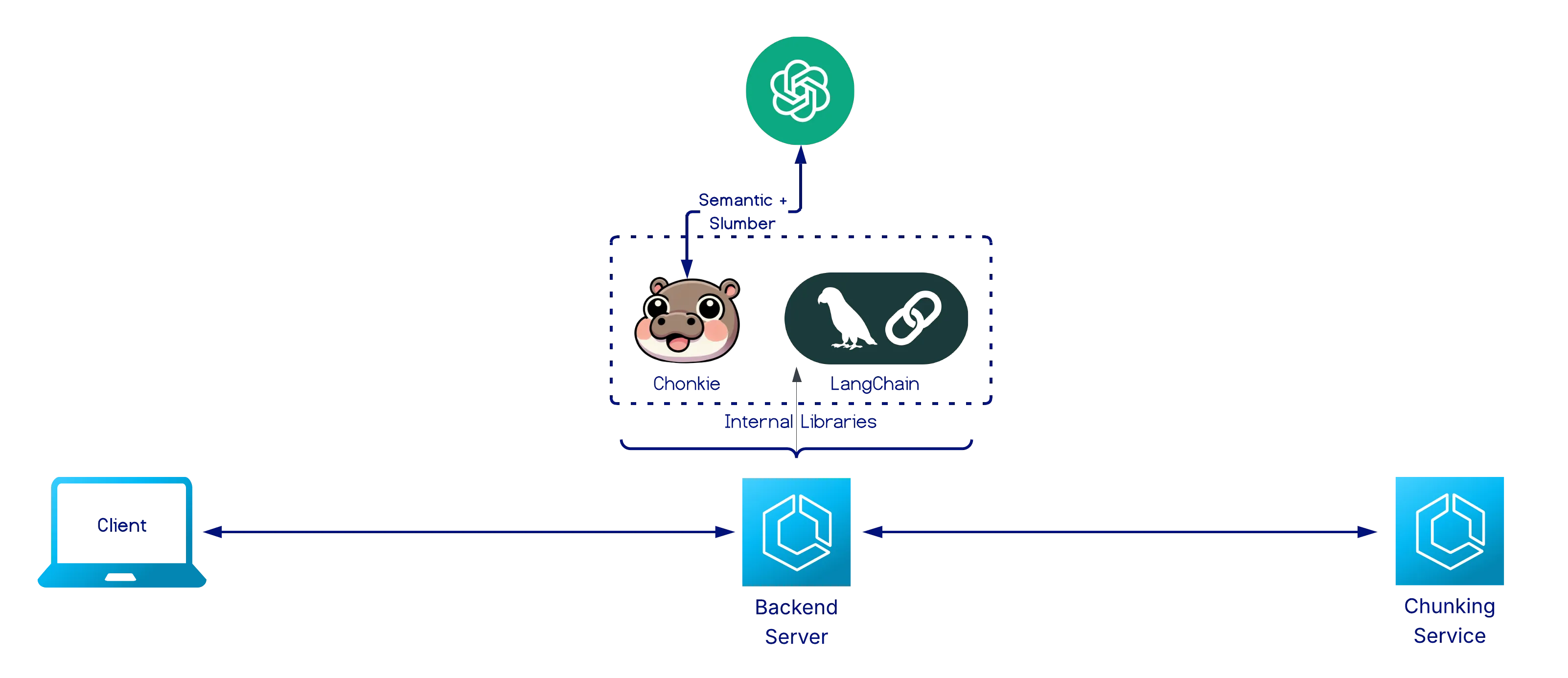
Functions as both a standalone chunking API and as an internal component supporting the visualization workflow. It provides a uniform interface for 8 chunkers from LangChain and Chonkie.
Given a chunking strategy and a document in the form of a string, the service:
- Instantiates the specified chunker.
- Executes chunking while abstracting over library-specific implementation differences.
- Returns structured chunk objects enriched with metadata (e.g., start/end indexes and token counts).
The service standardizes chunk output across different chunker providers, enabling consistent visualization and comparison.
Evaluation Service
Section titled “Evaluation Service”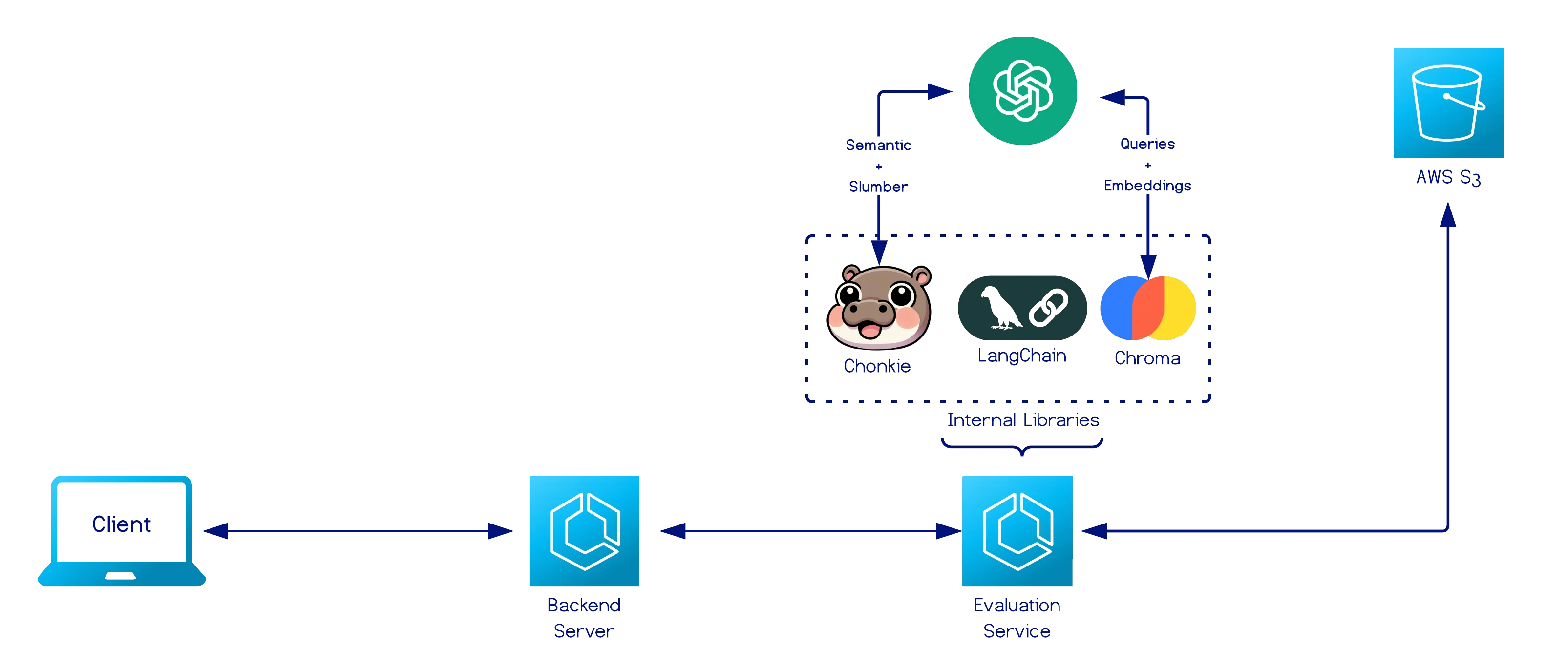
Operates as both a self-contained chunking-evaluation API and an internal microservice powering retrieval-based assessment in the platform.
Given an evaluation request that contains a chunking strategy and a document identifier, the service:
- Fetches the document from an S3 bucket via the document id.
- Applies the specified chunking strategy to generate candidate chunks.
- Searches for associated reference data (queries + ground-truth excerpts) sharing the same document id in the same bucket.
- If found, the reference set is reused.
- If not found, the service generates it using an LLM and stores it as a CSV file in S3.
- Computes four retrieval-based metrics and returns them along with the path to the generated or reused reference file.
Backend Server
Section titled “Backend Server”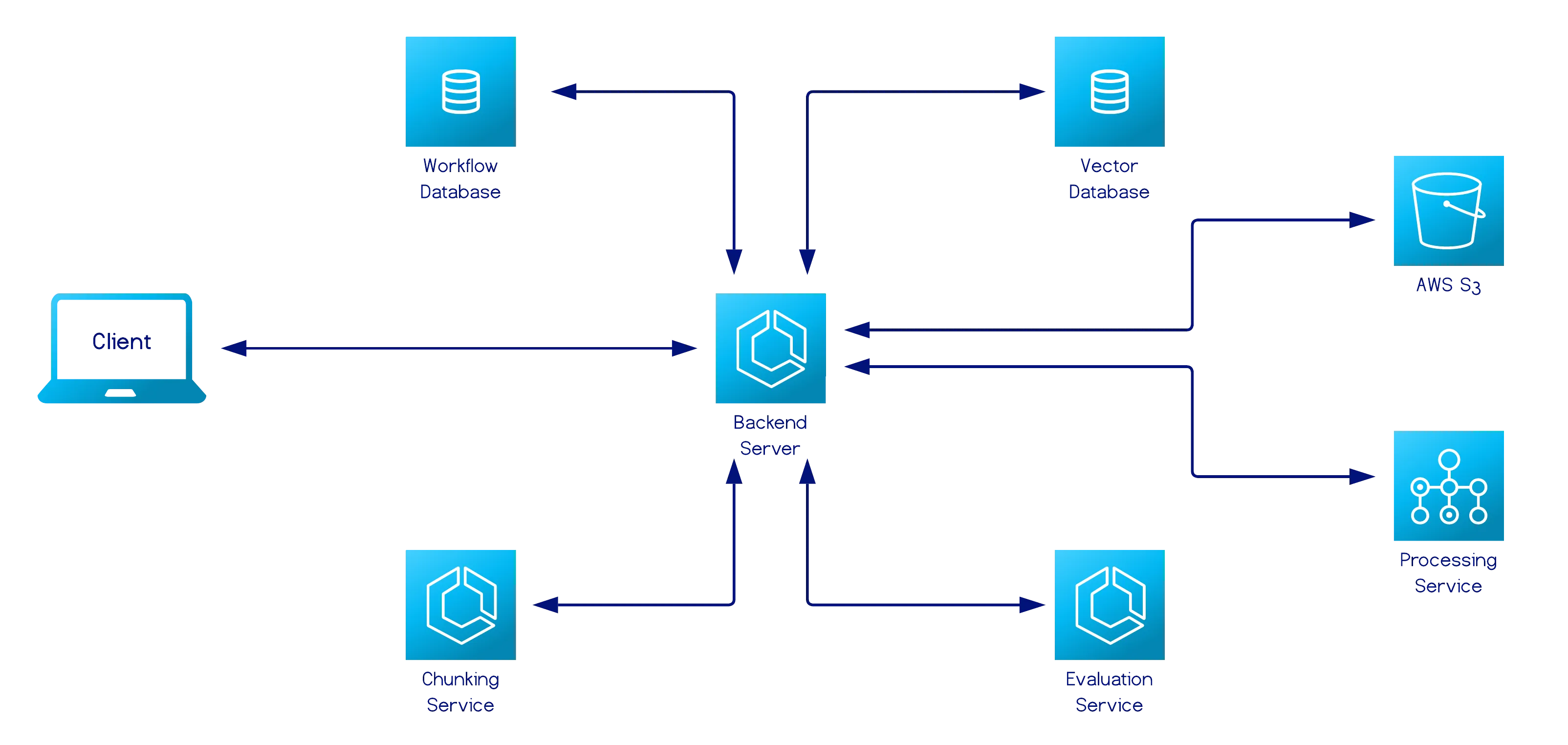
Orchestrates all user-facing workflows.
Its responsibilities include:
- Exposing a REST API for all client interactions, which provides predictable, stateless endpoints that map directly to user actions.
- Managing document lifecycle operations, such as uploading, normalizing, and reading.
- Forwarding requests to the chunking and evaluation services, using shared Pydantic models across all three services to ensure type-safe communication.
- Performing lightweight auxiliary operations that support visualization and experimentation.
- Persisting workflow metadata and results in the evaluation database.
This service acts as the central coordinator across all experimentation workflows and supports the RAG data ingestion pipeline.
Client
Section titled “Client”Provides the user interface for configuring, executing, and comparing chunking experimentation workflows, as well as deploying ingestion pipelines for downstream systems and workflows. Its responsibilities include:
- Allowing the user to upload sample documents and configure chunking strategies.
- Rendering the interactive visualization of the resulting chunks and their metadata, along with summary statistics.
- Triggering retrieval-based evaluation of a specified chunking strategy and presenting evaluation metrics.
- Enabling side-by-side comparison of the results of multiple experimentation workflows.
- Supporting deployment of ingestion workflows by connecting to an S3 data source and initiating a managed ingestion pipeline.
- Providing real-time status updates for ingestion workflows.
Data Ingestion Pipeline
Section titled “Data Ingestion Pipeline”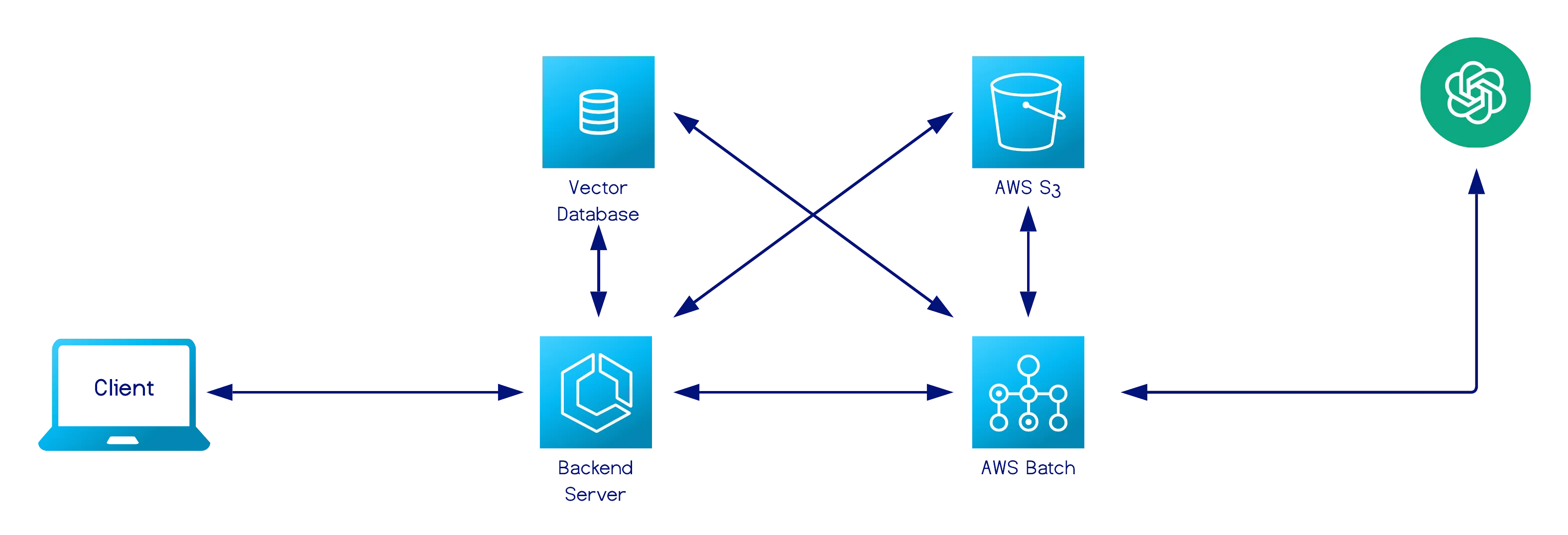
The ingestion pipeline is the production component of Chunkwise, responsible for applying a finalized chunking strategy to the user’s entire corpus and storing chunk embeddings for downstream retrieval use cases. When a user deploys a workflow, the backend initiates a processing sequence that operates across all documents associated with the knowledge base.

The ingestion pipeline performs four responsibilities:
- Retrieve and normalize documents - It reads document contents from storage and applies the same normalization used in the experimentation platform to ensure consistent, tokenizer-safe text.
- Apply the validated chunking strategy - Each document is chunked using the exact configuration the user selected during experimentation.
- Generate embeddings for each chunk - The pipeline computes vector embeddings for all chunks to prepare them for retrieval.
- Write chunks and embeddings to the vector database - The resulting chunk metadata and embedding vectors are stored in a structured table that is dedicated to the deployed workflow.
End-to-End Workflow
Section titled “End-to-End Workflow”Below is the end-to-end lifecycle of a Chunkwise workflow, which begins with file upload and processing, proceeds through chunk visualization and retrieval-based evaluation, and culminates in ingesting a user-provided dataset into a vector store using a specified chunking strategy.
1. Workflow Creation
The user creates a new workflow from the UI. The backend creates a corresponding workflow record in the evaluation database (an RDS Postgres instance).
2. Document Handling
The backend retrieves a list of previously uploaded documents from the private S3 bucket provisioned during deployment. The user may select an existing document or upload a new .txt or .md file (limited to 50 KB for latency considerations). The backend then normalizes the document by converting Unicode characters (e.g., smart quotes and dashes) to ASCII characters, preparing it for safe consumption by downstream services.
3. Chunk Visualization
Chunk visualization requires three inputs:
- A sample document
- A chunker
- Chunking parameters
As the user adjusts any of these inputs, the backend:
- Updates the workflow record.
- Invokes the chunking service with the selected chunking strategy and document.
- Receives chunks with metadata and returns them to the client for interactive visualization and statistics computation.
This provides real-time feedback as the user iterates on different chunking strategies.
4. Chunking Evaluation
When the user triggers evaluation on the selected chunking strategy, the service:
- Loads the document from S3.
- Retrieves or generates the associated reference set (queries + excerpts).
- Computes the four retrieval-based metrics.
- Returns the evaluation results to the backend.
The backend extracts the metrics from the evaluation response and stores them in the workflow record. It then displays them in the UI.
5. Workflow Comparison
The comparison view allows the user to analyze how different chunking strategies behave across workflows by holding selected parameters constant while changing others. This enables more systematic and controlled evaluation of chunking quality.
6. Deployment to Ingestion Pipeline
When a user deploys a selected chunking strategy, the backend triggers the ingestion pipeline. The pipeline processes the entire corpus using the finalized strategy and stores chunks and embeddings in the pipeline database for downstream retrieval systems.
Deployment Architecture (AWS)
Section titled “Deployment Architecture (AWS)”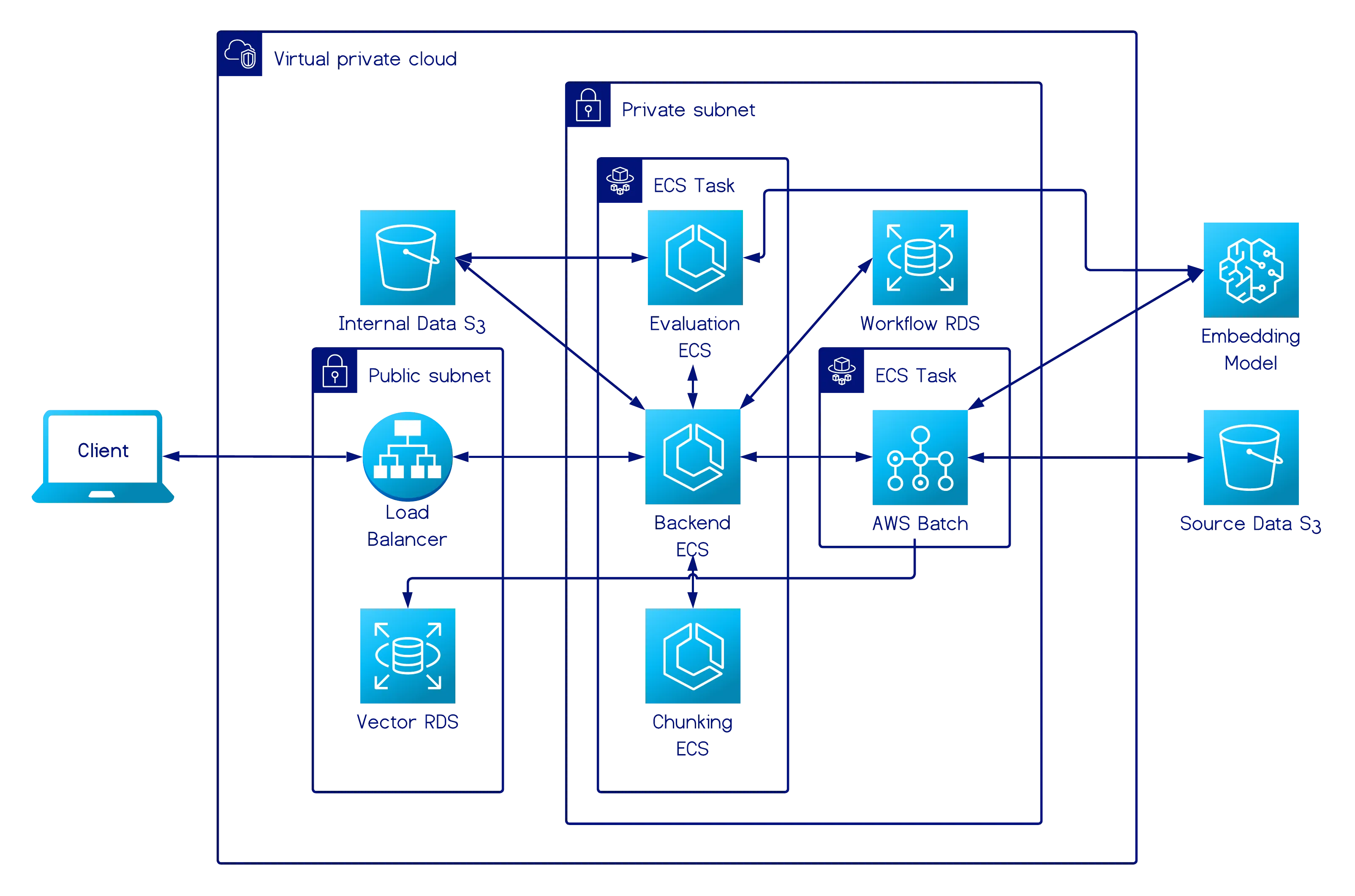
Platform Architecture (Core Services)
Section titled “Platform Architecture (Core Services)”We deployed the experimentation platform on AWS ECS Fargate, containerizing all three microservices—backend, chunking, and evaluation—and running them within a single ECS cluster for operational simplicity. Only the backend service is internet-facing and exposed publicly through an Application Load Balancer, so that the locally running client can connect securely to the platform. The chunking and evaluation services remain internal and communicate with the backend via AWS Cloud Map service discovery. The backend also interfaces with AWS Batch to initiate ingestion jobs when users deploy their corpus for processing.
The data layer consists of two core components:
- A private Amazon S3 bucket that stores uploaded documents and generated reference datasets.
- Two Amazon RDS PostgreSQL instances, which handle data persistence for the experimentation platform and the data ingestion pipeline, respectively.
We deployed two separate RDS instances because they serve different scopes and operational purposes. The experimentation RDS stores experimentation workflow records, including visualization HTML, chunk statistics, evaluation metrics, and metadata. It is deployed in private subnets and accessible only to internal services, since it supports developer-driven experimentation and does not participate in production workflows.
In contrast, the pipeline RDS is designed as a vector database, enabled with the pgvector extension, and stores chunks, embeddings, and document metadata for use in downstream retrieval workloads. This database is deployed in public subnets and intentionally kept publicly accessible to allow integration with external RAG systems, agentic workflows, and user-managed vector databases. The separation also ensures that experimentation workloads, which are often iterative and bursty, cannot affect production retrieval performance. It also allows each subsystem to scale, secure, and evolve independently according to its individual needs.
Within the overall Chunkwise architecture, only the backend service communicates with the two RDS instances, while all other services remain stateless. This architecture separates compute, storage, and networking concerns, keeps all internal traffic inside the VPC, and provides autoscaling, resilience, and isolation for each microservice with minimal operational overhead.
Data Ingestion Pipeline Architecture (Batch Processing Jobs)
Section titled “Data Ingestion Pipeline Architecture (Batch Processing Jobs)”We used AWS Batch with Fargate to handle our AI-focused ETL pipeline. When the server receives a deploy request, it retrieves the key and size for each document in the user’s S3 bucket and evenly distributes them among four jobs based on size. Jobs are created according to the job definition’s specifications (1 vCPU and 8 GB of memory) and placed in the job queue. Each job retrieves its documents from S3, chunks and embeds them, and stores the vector embeddings and metadata in RDS.